
White Papers
What Is Next-Generation Sequencing? How Does It Work? And Why Does It Matter for Food Safety?
Introduction
Next-generation sequencing (NGS) is changing the food industry. Since it is a relatively new technology, there are many misconceptions about NGS. For example, NGS is often mistaken for whole-genome sequencing (WGS), when, in actuality, sequencing an entire genome is only one application of NGS.
Food manufacturers and service labs can use NGS to do so much more, including routine pathogen testing, persistent/transient analysis, GMO verification, authenticity testing, and microbiome studies.
Though NGS is gaining popularity within the food industry, it can be difficult to find useful resources to learn about this food safety technology. So, we’ve compiled a quick tutorial for you on next-generation sequencing, including links to useful articles and videos.
Whether you want to brush up on the basics of NGS, need help with the scientific details of next-generation sequencing, or are simply curious about its real-world applications, this is the page for you.
To jump to a specific section, use these links:
- So, What Is Sequencing?
- How Does First-Generation Sequencing Work?
- How Does Next-Generation Sequencing Work?
- How Does Nanopore Sequencing Work?
- How Do You Interpret the Data?
- How Does Next-Generation Sequencing Fit in Food Safety?
- Glossary of Terms
Note: We will dive into the technical details of sequencing. If you’re looking for a more general introduction to NGS and the technology’s benefits for your business, check out this resource: An Introduction to Next-Generation Sequencing for Food Safety Labs.
So, What Is Sequencing?
Let’s take a quick journey back in time. In 1953, James Watson and Francis Crick discovered the structure of DNA. Specifically, they determined that DNA has a double-helix structure, which is composed of sugar (deoxyribose), phosphorous, and nitrogenous bases (adenine, guanine, cytosine, and thymine). In many ways, their contribution provided the key to understanding how genetic material is passed on, and it paved the way for sequencing.
When scientists refer to sequencing, they are referring to the sequence in which the bases appear, that is, the order in which adenine, guanine, cytosine, and thymine appear. But instead of referring to the bases by name, they use letters: A, G, C, and T. And as you might recall from biology class, As bind with Ts, and Cs bind with Gs.
When scientists started sequencing genomes, they relied on what we call today “first-generation sequencing.” The most common technique was developed in 1977 by Frederick Sanger, and it was used for several decades. (Walter Gilbert invented his own method, but it didn’t catch on.)
While Sanger sequencing enabled scientists to read significant sections of the genome fairly accurately, there were a few problems. Chief among them were cost, turnaround time, and a lack of sequence redundancy. Sanger sequencing was relatively expensive and time-consuming since the technology was limited in the number of sequences it could read simultaneously. First-generation sequencing generated only one sequence per genome. Imagine what would happen if there were an error at some point. The machine wouldn’t create additional (i.e. redundant) sequences.
NGS, on the other hand, can generate and interpret millions of sequences simultaneously. In other words, massive amounts of data are processed in a far shorter period of time and at a lower cost. As a result, NGS methods generate redundancies in reads, meaning they will look at the same region of nucleotides multiple times, thereby reducing the likelihood of error. This is often referred to in the NGS literature as Sequencing depth or coverage.
To further illustrate the shortcomings of first-generation sequencing, let’s turn to the classic example of the Human Genome Project. Started in 1990, the project sequenced the entire human genome (approximately 3 billion base pairs long) using first-generation sequencing. It took almost 15 years, required the cooperation of several labs around the world, and cost approximately $2.7B USD.
Nowadays, NGS sequencers have the potential to quickly and reliably sequence the entire human genome for less than $1,000 USD. In other words, NGS technologies can do the work of hundreds of Sanger machines for a fraction of the cost and in a fraction of the time.
So, what’s the difference? What makes NGS sequencers so much quicker and cost-effective? To understand that, let’s dive into how sequencing technologies work.
How Does First-Generation Sequencing Work?
As we mentioned above, Frederick Sanger developed a form of first-generation sequencing in the 1970s, and it was the most common form of sequencing well into the early aughts. (454 introduced the first commercial NGS instrument in 2005.)
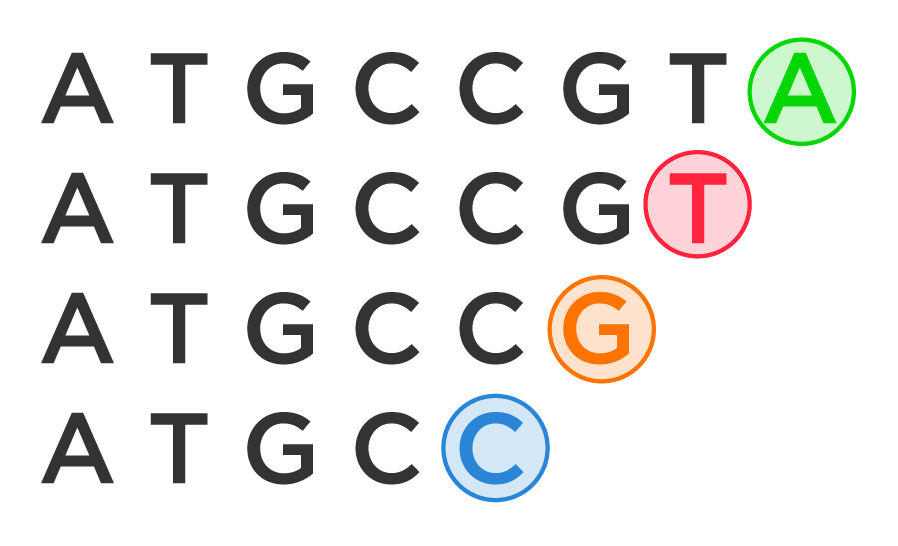
In a nutshell, Sanger sequencing involves making many copies of a target region of DNA. Each copy will be of a different length, and the technology will use the length of the copied strand to read the order of the bases.
For example, let’s say one copy might read ATGCC. Another copy might read ATGCCG, another ATGCCGT, and another ATGCCGTA. The sequencing technology will look at the last letter on the shortest strand and say, “Aha! The first letter is C.” Then, it will look at the last letter on the second shortest strand and say, “Aha! The next letter is G.” And so on.
But how does the first-generation sequencing technology do that? Sanger sequencing relies on the following components:
- A DNA template: This is a strand from a specific region of a DNA molecule that scientists want to sequence (i.e. understand the order in which the As, Cs, Gs, and Ts appear).
- A primer: This is a short piece of complementary single-stranded DNA that attaches to the DNA template. It is crucial for making copies of the DNA template.
- A DNA polymerase enzyme: This helps the primer attach to the template and synthesizes DNA formation.
- The four DNA nucleotides: Deoxyadenosine triphosphate (dATP or A for short), Deoxyguanosine triphosphate (dGTP, or G for short), Deoxycytidine triphosphate (dCTP or C for short), and Thymidine triphosphate (dTTP, or T for short).
- Dideoxy versions of all four nucleotides: ddATP, ddTTP, ddCTP, ddGTP. As the DNA template is sequenced, the dideoxynucleotides attach themselves to the end of the replicated DNA and indicate to the DNA polymerase, “Hey, this chain of DNA is done. It’s time to start a new chain.” Each dideoxynucleotide is labeled with one of four colored dyes, and the colors are used to read the order of the nucleotides.
Here’s a helpful video that helps you visualize how Sanger sequencing works.

To recap, the DNA to be sequenced is combined with primers, DNA polymerase, and DNA nucleotides. The dye-labeled, chain-terminating dideoxynucleotides are added as well, but there are fewer of them.
The mixture is first heated to denature (separate) the DNA, revealing a single-stranded template. Then, it’s cooled to a temperature at which the primer can attach to the DNA template. Once the primer has attached, the temperature is raised again, allowing the DNA polymerase to synthesize new DNA starting from the primer. The DNA polymerase will continue adding nucleotides to the chain until a dideoxynucleotide is added, signaling to the DNA polymerase enzyme, “Hey, this chain is done. Let’s start a new one.”
The process is repeated over and over until there is a dideoxynucleotide at every single position of the target DNA beyond the length of the primer. The end result: A bunch of DNA fragments, each of a different length, each with an A, C, G, or T at the end. (Technically, each fragment ends with a ddA, ddC, ddG, or ddT because each fragment ends with a dideoxynucleotide.) At this point, though, we don’t know the order in which those As, Cs, Gs, and Ts appear.
That’s where capillary gel electrophoresis enters the picture. Essentially, the gel is a thin gelatin-like medium used to separate DNA by size. Since DNA is charged, an electric current draws the DNA fragments through the gel; the shorter fragments move quickly through the pores of the gel, while the long fragments move more slowly.
As each fragment moves through the end of the tube, a laser illuminates it, and the dye color from the dideoxynucleotide is registered on the detector. A chromatogram is produced, based on a series of peaks in fluorescence intensity, and the DNA sequence is derived from the peaks in the chromatogram.
The Limitations of Sanger Sequencing
In its day, Sanger sequencing was a huge breakthrough, but time has shown the technology’s limitations. Compared to NGS, Sanger sequencing remains extremely costly. In 2011, it cost as much as $500 per Mb, which means that the human genome would cost about $1.5M to sequence. At the time, some other NGS technologies were priced at $0.10 per Mb.
(Side note: Mb stands for megabase. 1 megabase is equal to 1,000,000 bases.)
In addition to the question of cost, there are some more technical issues. For example, Sanger methods can only sequence short pieces of DNA, typically between 300 and 1,000 base pairs. Moreover, sequence quality often degrades after 700 to 900 base pairs.
You can probably see why another generation of sequencing was needed…
How Does Next-Generation Sequencing Work?
An important preface: There isn’t a singular NGS technique that everyone uses. Instead, NGS refers to a variety of methods that enable researchers and professionals to generate and interpret millions of sequences at the same time.
A number of NGS technologies, such as Illumina’s reversible dye-terminator sequencing, Pacific Biosciences’ single-molecule real-time sequencing, and Ion Torrent semiconductor sequencing, are based on the sequencing-by-synthesis principle.
But there are other methods of next-generation sequencing. One of the earliest NGS technologies was 454 (later acquired by Roche), which used pyrosequencing. In addition, there is sequencing-by-ligation. We won’t dive into each of those techniques, but you can learn more about pyrosequencing, sequencing-as-synthesis, and sequencing-by-ligation by watching this video.
For the purposes of explanation, let’s discuss Illumina’s sequencing-by-synthesis because we can easily compare and contrast it with Sanger sequencing.
Just as we saw in Sanger sequencing, sequencing-by-synthesis has similar key components:
- A DNA template
- A primer
- A DNA polymerase enzyme
- The four DNA nucleotides (dATP, dTTP, dCTP, dGTP)
- Dideoxy versions of all four nucleotides (ddATP, ddTTP, ddCTP, ddGTP)
But one key difference lies in the number of templates. Whereas Sanger sequencing starts with one template, NGS techniques can start with millions of templates.
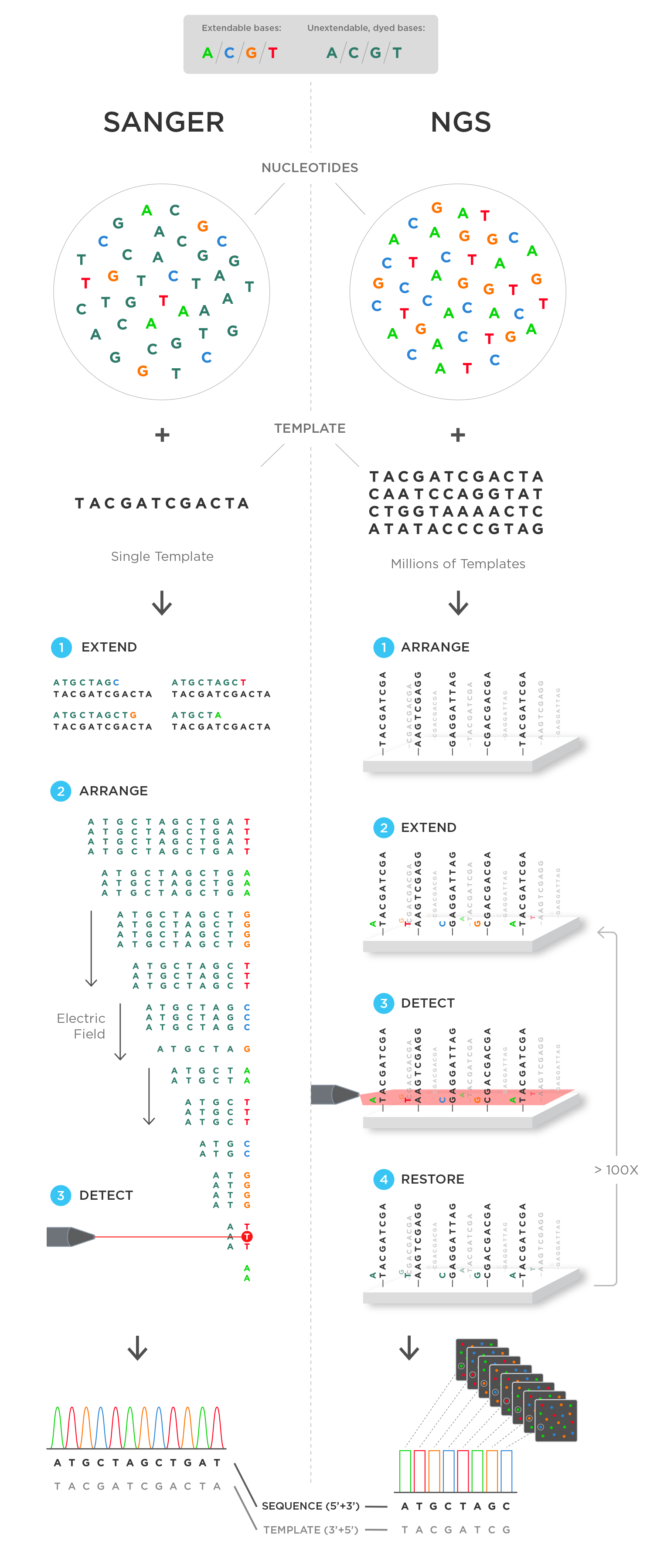
In NGS, the millions of template strands are attached to an immovable structure. Often times, it is a glass slide, but some technologies use beads. Whatever it may be, it has to be immovable because the template strands remain fixed at the same position throughout the entire sequencing process.
Once the strands are in place, the next step is to extend the primers. Each template is extended by a single chain-terminating, non-extendable dideoxynucleotide. Then, a microscope captures both the position of each template and the fluorescent color and intensity of the dideoxynucleotides. As we saw in Sanger sequencing, each color is associated with a different nucleotide.
What happens next is quite different from Sanger sequencing. A restoration process occurs. The chain-terminating, non-extendable dideoxynucleotides are replaced with regular, extendable nucleotides. So, a ddA is replaced by a A, a ddC by a C, and so on. This allows the templates to undergo subsequent rounds of single-based extension and imaging.
So, let’s take a simple example. For the sake of simplicity, let’s say that we have a really short template: TACGGCAT. The template’s complementary strand would read ATGCCGTA. And let’s say that we have a primer that reads ATGC.
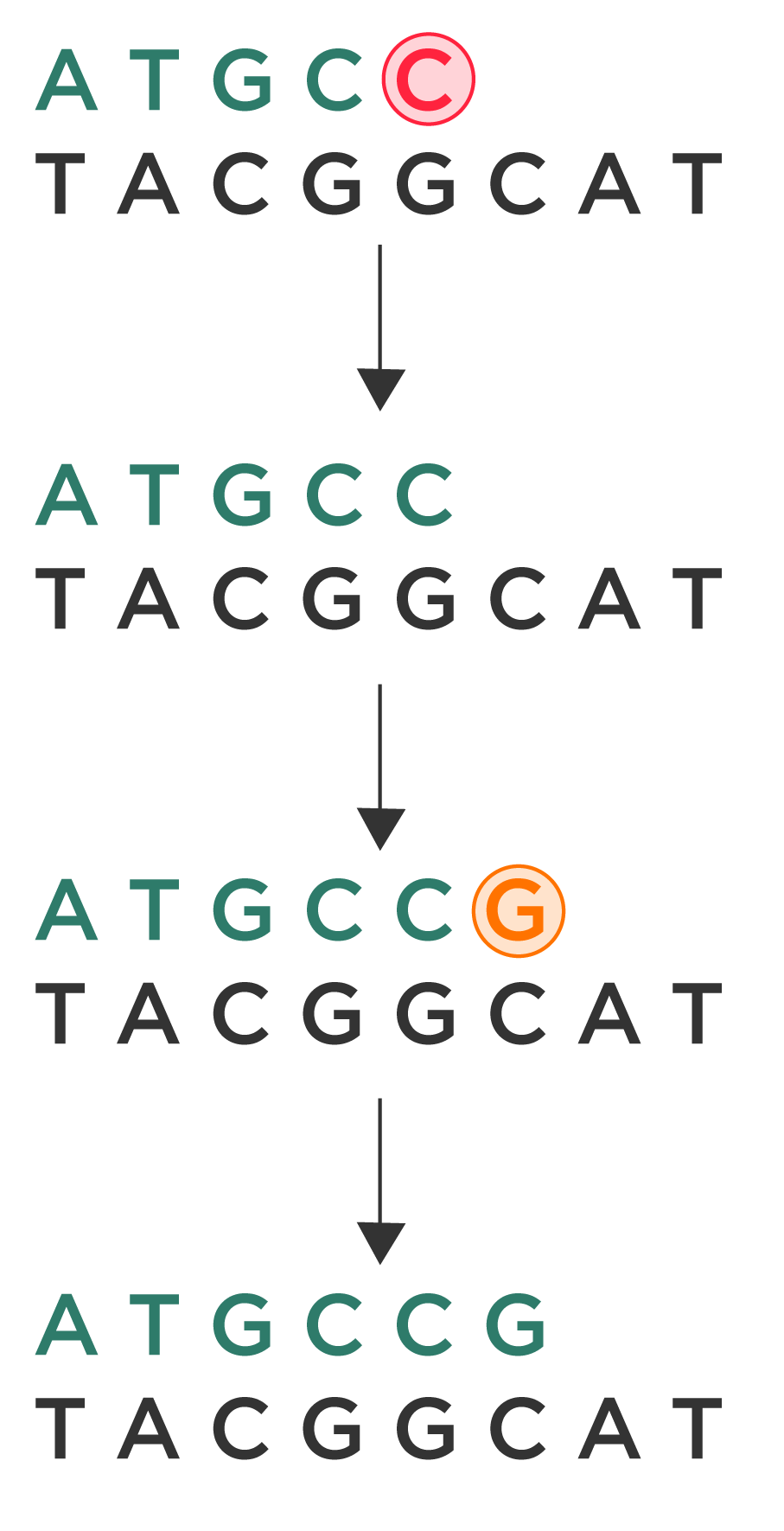
During the first round of extension, a deoxynucleotide would be attached to the primer. So, the complementary strand now reads “ATGCddC,” with a chain-terminating, non-extendable ddC at the end. A snapshot would be taken of this extended primer (along with the millions of other primers that are being extended simultaneously). Then, the chain-terminating, non-extendable ddC at the end would be replaced with a regular, extendable C nucleotide: ATGCC.
During the next round of extension, a ddG deoxynucleotide would be added to the complementary strand, then imaged, and finally replaced with a regular, extendable G nucleotide. And so on.
How Does Nanopore Sequencing Work?
In the previous section, we mentioned several methods of NGS. One of the most recent developments has been nanopore sequencing. Some call it third-generation sequencing. Others call it fourth-generation sequencing. Regardless, nanopore sequencing represents a completely different method of sequencing. (It is also the sequencing technique that Clear Safety, our flagship product, uses.)
David Wilson Deamer, Dan Branton, and George Church first proposed nanopore sequencing in the 1990s. Their vision is fairly simple to understand. Snake DNA molecules through a miniscule opening called a nanopore. Run a small voltage across the opening. As the nucleotide chain of the DNA molecule passes through the pore, each nucleotide blocks the pore momentarily and impedes the current through the pore in a unique way, thus producing a unique signal profile. Deduce a DNA strand’s sequence based on the series of these signal profiles.
Fast forward several years, and Deamer’s, Branton’s, and Church’s visions have been commercialized. Today, there are many ways to produce ionic currents for DNA sequencing. Some technologies use protein nanopores while others use graphene or other solid-state nanopores. Regardless of the type of nanopore, the method remains essentially the same.
First, the DNA strand first passes through an enzyme that unwinds the DNA and feeds one strand through a nanopore.
A small (≈100 mV) voltage bias is imposed across the pore. Inside the pore are aqueous electrolytes. (Quick chemistry reminder: When a solute dissolves in water to form a charged atom, also known as an ion, it is called an electrolyte.) The resulting ionic current through the pore is measured, and as the DNA strand passes through the pore, each nucleotide disrupts the electrical current differently, producing a readout of the base sequence.
All of this happens quite quickly, given that nanopore sequencing can handle 450 bases per second.
To help you visualize how this works, watch Oxford Nanopore’s explainer video.
The Advantages of Nanopore Sequencing
As we mentioned above, nanopore sequencing technology is quite quick, handling 450 bases per second and resulting in faster turnaround times and lower costs. In fact, nanopore sequencing has made routine pathogen testing as fast as and as inexpensive as PCR testing.
When nanopore sequencing was first developed, there was concern that the technology’s speed would adversely affect its accuracy. This was primarily a concern for researchers who were doing de novo sequencing. (De novo sequencing refers to the sequencing of a novel genome that does not have a reference genome. During this process, nothing is known about the genome, and the entire genome must be sequenced.) However, recent developments have improved the accuracy of nanopore sequencing, even for de novo sequencing.
In the food safety lab, de novo sequencing is not a major concern. Routine pathogen testing, for example, always uses a reference genome. Moreover, it relies on targeted sequencing, which means that only certain regions of the genome are sequenced. As a result, shorter pieces of genetic material are analyzed at one time, and each of those pieces are analyzed multiple times. We will discuss targeted sequencing below.
How Do You Interpret the Data?
DNA sequencing can produce millions of data points. No human being can manually process all the strings of As, Ts, Cs, and Gs, so scientists must rely on bioinformatics procedures to interpret the data.
As a field, bioinformatics has its roots in comparing sequences to one another. Through comparison, we can make inferences about new sequences based on their similarity with previously characterized sequences. The process relies on powerful algorithms that are often used in the context of natural language processing. (If you think of it, DNA sequences are strings of letters that make up a language in their own right.)
Here’s how it works.
After a sequencing run, we need to compare the sequences to existing sequences. This means that we need a carefully curated database of sequences that have previously been identified and annotated. That way, we can easily compare the sequencing data from test runs to known sequences. In turn, we can use the known sequences in our database to identify the pathogens or other microorganisms in a sample.
This is not unlike looking up an unfamiliar word in a dictionary to learn its meaning, but instead of words like “Salmonella,” we have strings of As, Ts, Cs, and Gs that are indicative of Salmonella.
Data Analysis Doesn’t Have to Be Hard
You might be wondering if you have to employ a team of bioinformaticians to use NGS in your food safety lab. Luckily, NGS platforms that are built for food safety testing have algorithms baked into the technology. That way, food safety professionals don’t have to do the heavy lifting when it comes to data analysis. Instead, they can log into their software and look at reports like the one below.
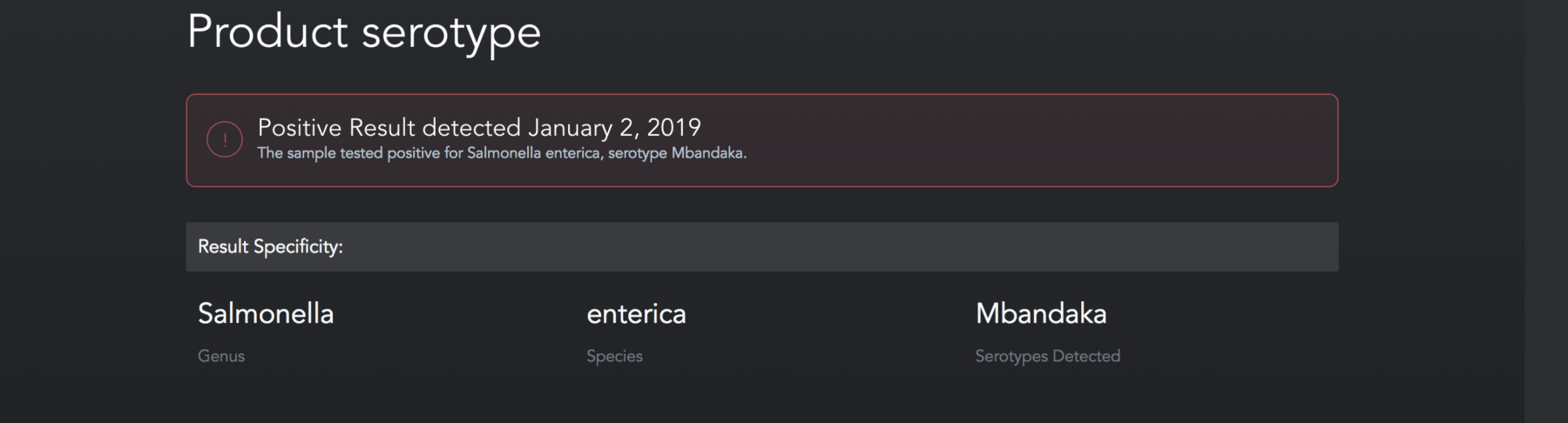
As you can see, this report determined which Salmonella serotype was found in each sample. To produce a serotype report like the one above, the platform looks at specific targeted regions of a sample’s DNA. Then, it sequences those regions. Next, it uses algorithms to compare the sample’s sequences to reference sequences of various Salmonella serovars. If the sequence of the sample matches the sequence of, say, Typhimurium, then, the sample comes back positive for Typhimurium.
A Note about Targeted Sequencing
Evolution has endowed bacterial genomes with uniquely identifiable stretches of DNA that not only provide bacteria with their characteristic biological functions, but also serve as signatures that make a particular bacterial genome distinguishable from the genomes of other bacteria. Within the genome, there are certain regions that are more telling than others.
By using a targeted sequencing approach, we are able to look at the idiosyncratic parts of a pathogen’s genome that are most informative for detection and subtyping.
In this way, we are able to maximize the information gleaned from sequencing without wasting time and computational power on noise. Think of it like a fishing expedition. What if you had bait that could attract only the most nutritious fish and you never had to worry about catching refuse on your line? That’s the way targeted sequencing works. You pull out the most information-rich sequence patterns from a sea of DNA present in a given food sample.
For use cases like routine pathogen testing or serotyping, a targeted approach is far superior to WGS because it is far cheaper and faster than WGS. More on this in the next section.
How Does Next-Generation Sequencing Fit in Food Safety?
NGS has already had a huge impact on food safety. It is being used for authenticity testing, traceback analyses, GMO validation, microbiome studies, and routine pathogen testing. In the articles below, you can find some of the most common applications, as well as some of the most common misconceptions.
How Next-Generation Sequencing Will Impact Your Food Safety Program
Think of NGS like a smartphone. Just as we have different applications on our smartphones, there are different applications of NGS. In this article, Sasan Amini explains why routine pathogen testing requires targeted NGS, microbiome studies require shotgun metagenomics, and why traceback analysis requires WGS.
Why Whole Genome Sequencing Is Not the Right Tool for Routine Pathogen Testing
Most food safety professionals have heard of whole-genome sequencing. Nevertheless, WGS isn’t always the right tool for the job. Sasan Amini explains why WGS shouldn’t be used for routine pathogen testing and why targeted sequencing should be used instead
Over the years, scientists have developed several methods to compare the DNA signatures of microorganisms. This resource compares pulsed-field gel electrophoresis (PFGE), ribotyping, Similarity Analysis through targeted sequencing, and whole-genome sequencing (WGS).
How Dafgård is Using NGS to Ensure Food Safety
The Swedish food provider has been able to leverage Clear Labs’ platform to use a single test to screen each batch of its food for allergens, missing ingredients, and even the unexpected – like an unintended ingredient or pathogen. Learn more in this blog post.
Hot Dog Study: Next-Generation Sequencing as a Novel Tool for Quality Control of Food Products
Have you ever wondered what is in your hot dogs? We purchased 345 hot dog products and used next-generation sequencing to determine whether your beef hot dog is really made of beef.
When you’re eating red snapper at a restaurant, are you really eating red snapper? We used NGS to find out.
Glossary of Terms
As you learn more about NGS, you’re bound to run across a few unfamiliar terms. So, here are a few of the most common words with their definitions.
Bioinformatics: In short, this is the process of collecting, analyzing, and interpreting sequencing data.
Coverage: This is the number of times that a particular target region of DNA is sequenced during a sequencing run. Sequencing reactions can be prone to errors, so typically, NGS technology requires 30x coverage, meaning each target region must be sequenced 30 times. When the depth of coverage exceeds 30x, then, we use the term deep sequencing.
Denature: In general, denaturation occurs when the shape of a protein is altered. In sequencing, denaturation often refers to splitting the double helix structure.
De Novo Sequencing: This refers to sequencing a novel genome where there is no reference sequence available.
Genome: The complete set of genes or genetic material in an organism.
GenomeTrakr: This is a network of laboratories that have used WGS for pathogen detection. According to the FDA website, the network regularly sequences over 9,000 isolates per month.
Library: This involves generating a collection of DNA fragments for sequencing. During the library preparation process, special adapters are ligated to the ends of the fragments. These adapters can help with attaching the fragments to solid surfaces, providing priming location for both amplification and sequencing primers, and providing barcoding for multiplexing different samples in the same run. Library preparation is required before each sequencing run.
Multiplexing: This allows large numbers of libraries (i.e. DNA fragments with adapters attached to them) to be pooled and sequenced simultaneously.
Reads: This is the output of a NGS sequencing reaction. A read is an uninterrupted series of nucleotides in a specific order.
Read Length: This is the number of base pairs that you can read at one time.
Shotgun Sequencing: DNA is randomly shredded into many small fragments that can be sequenced individually. Then, the sequences of these fragments are reassembled into their order, ultimately producing the complete sequence.
Shotgun Metagenomics: This is a specific type of shotgun sequencing. If the sample is complex (i.e. has multiple populations) as most environmental samples do, then applying shotgun sequencing generates shotgun metagenomics data, which can be used to identify multiple microorganisms in a sample.
Targeted Sequencing: This is a specific application of next-generation sequencing. Instead of reading the entire genome, it looks at specific regions that are idiosyncratic and useful for identifying a microorganism’s identity.
WGS: Whole-genome sequencing. WGS uses NGS platforms to look at the entire DNA of an organism. It is non-targeted, which means it is not necessary to know in advance what is being detected. In WGS, the entire genome is cut into small regions, with adaptors attached to the fragments to sequence each piece in both directions. The generated sequences are then assembled into single long pieces of the whole genome. WGS produces sequences 30 times the size of the genome, providing redundancy that allows for a deeper analysis.
What’s Next?
NGS is rapidly changing the food safety industry. We will keep this list of NGS resources up to date so that you can stay current.
Do you have a useful NGS resource you want us to add to our list? Did we miss anything? Shoot us an email at inquiries@clearlabs.com.